Description
The coronavirus disease 2019 (COVID-19), caused by the SARS-CoV-2 virus, has emerged as one of the most widespread and devastating pandemics in the history of humanity. To date, despite the availability of vaccines, the pandemic still cannot be fully controlled due to rapid mutation of the virus. Thus, the development of safe and effective therapeutic options for COVID-19 is still an unmet need.
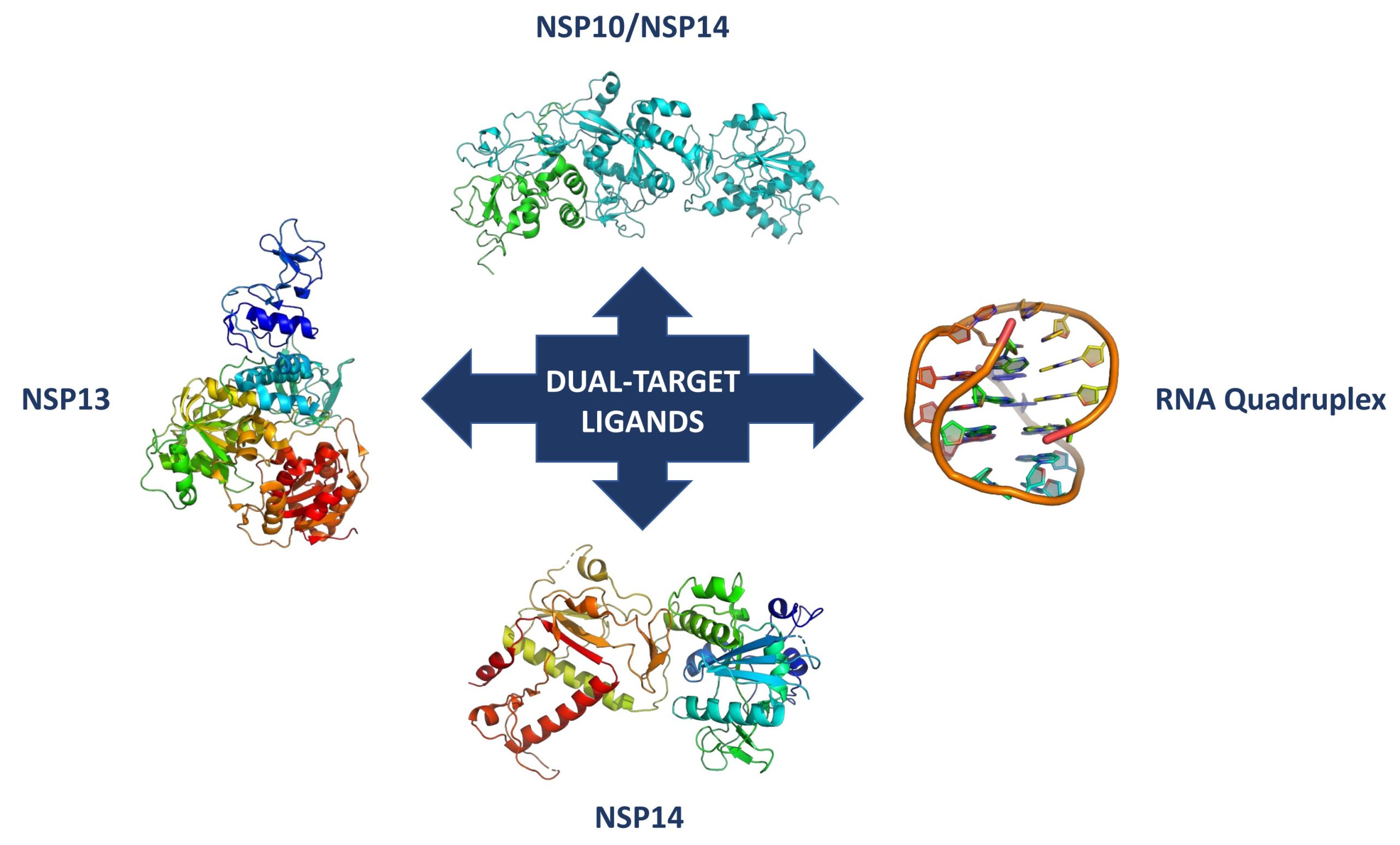
The MUTASAR project proposes a new therapeutic strategy to block the infection and progression of SARS-CoV-2 by using a multi-target approach, which involves either the simultaneous inhibition of two non-structural viral proteins, namely NSP13, NSP10, and NSP14 or the inhibition of one of these proteins and the targeting of viral RNA G-quadruplexes (G4s). All these targets have been demonstrated to play a key role in the viral replication. In particular, the helicase NSP13 is a vital enzyme, which unwinds RNA secondary structure within the 5′-untranslated region of the viral genome. On the other side, the bifunctional protein NSP14 consists of two domains: an N-terminal exoribonuclease (ExoN) domain, which plays a proofreading role for preventing lethal mutagenesis and a C-terminal domain, which functions as a (guanine-N7) methyl transferase (N7-MTase). Finally, NSP10 binds to NSP14 ExoN domain and is essential for the stimulation of its activity. SARS-CoV-2 virus is also characterized by alternative nucleic acid structures, like G-quadruplexes which play critical roles in several viruses, and some G4-targeting compounds have shown antiviral activity.
Two different approaches will be used in MUTASAR project: in the first case, in silico ligand-based and structure-based approaches will be utilized to identify inhibitors/ligands of the targets under investigation and then the shared hits, able to well recognize two of the four viral targets, will be selected. In the second case, multitarget ligands will be obtained by combining two single-target agents together through suitable linkers.
Objectives and expected results
The MUTASAR project will be realized by using a multidisciplinary approach, covering medicinal and computational chemistry, molecular, structural and chemical biology, and organic synthesis. In particular, the IBB unit will be involved in performing binding affinity measurements, inhibition assays and crystallographic studies. More in detail, the viral targets NSP10, NSP13, NSP14 will be cloned, expressed and purified in E. coli. The small molecules, identified and synthesized by the other two partners of the project, will be evaluated for their affinity against these targets and viral RNA G4s by means of several biophysical techniques such as microscale thermophoresis and/or fluorescence spectroscopy. Moreover, isothermal titration calorimetry, will be used to determine also enthalpy changes and binding stoichiometry of the interaction between the synthesized ligands and both the target proteins and the G4 viral RNA.
The best ligands emerged from these evaluations will be co-crystallized with the target protein/RNA in order to understand the molecular details responsible for the interaction and to provide a structural basis for further optimization.
Together with the other partners, IBB unit will be involved into the dissemination of the project results and it will coordinate the management of the project.
List of deliverables
- Dissociation and inhibition constants of the synthesized compounds against their viral target.
- Determination of the binding stoichiometry and the thermodynamic parameters related to the association processes.
- Crystal structure of viral target/ligand complex.
Project proponents
- Istituto di Biostrutture e Bioimmagini-CNR
Involved Entities
- Università degli studi di Napoli “Federico II”
- Università “Magna Grecia” di Catanzaro

